Better cellulases: lignocellulose to sugars
Cellulose is abundant, but woefully underutilized as a source of renewable fuels and chemicals. Cellulases decompose cellulose into its component sugar molecules, which in turn can be converted by microbes into valuable products. The problem is cost—the natural enzymes are slow and therefore too expensive for low-value products like fuels and commodity chemicals. We are trying to create better cellulases using a suite of approaches, including recombination and random mutagenesis. We have already discovered cellulases that are highly thermostable and significantly more active at elevated temperatures. This project lets us develop and test new protein engineering approaches in the context of a highly relevant application.
Here are just a few of our recent papers:
"A Structural Study of Hypocrea jecorina Cel5A," T. M. Lee., M. F. Farrow, F. H. Arnold, S. L. Mayo. Protein Science 20, 1935-1940 (2011).
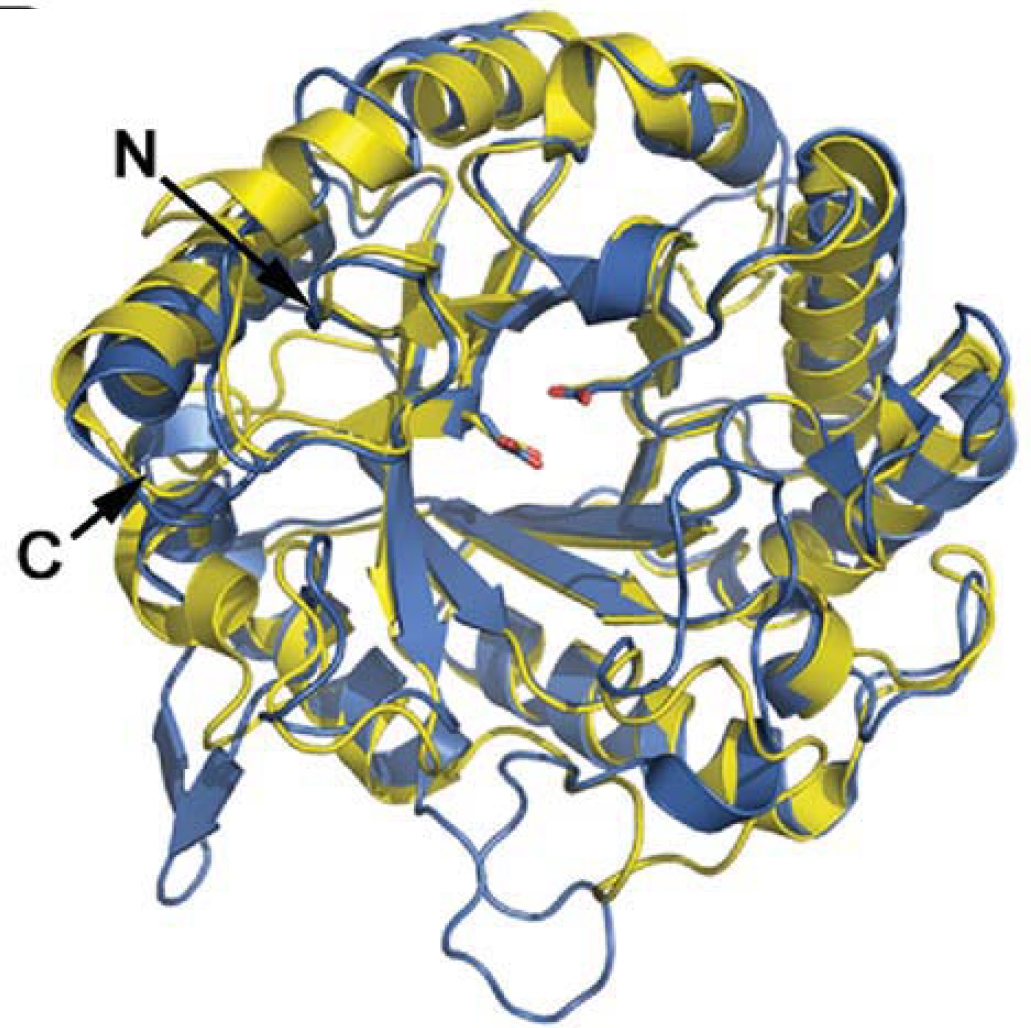 This paper reports the crystallization and structural determination of Hypocrea jecorina Cel5A. This enzyme is the major endoglucanase used in industrial hydrolysis of lignocellulase biomass. Structural efforts such as this help rational and semi-rational design of improved endoglucanases.
This paper reports the crystallization and structural determination of Hypocrea jecorina Cel5A. This enzyme is the major endoglucanase used in industrial hydrolysis of lignocellulase biomass. Structural efforts such as this help rational and semi-rational design of improved endoglucanases.
"Comparison of Family 9 Cellulases from Mesophilic and Thermophilic Bacteria,"F. Mingardon, J. D. Bagert, C. Maisonnier, D. L. Trudeau, F. H. Arnold. Applied and Environmental Microbiology 77(4), 1436-1442 (2011).
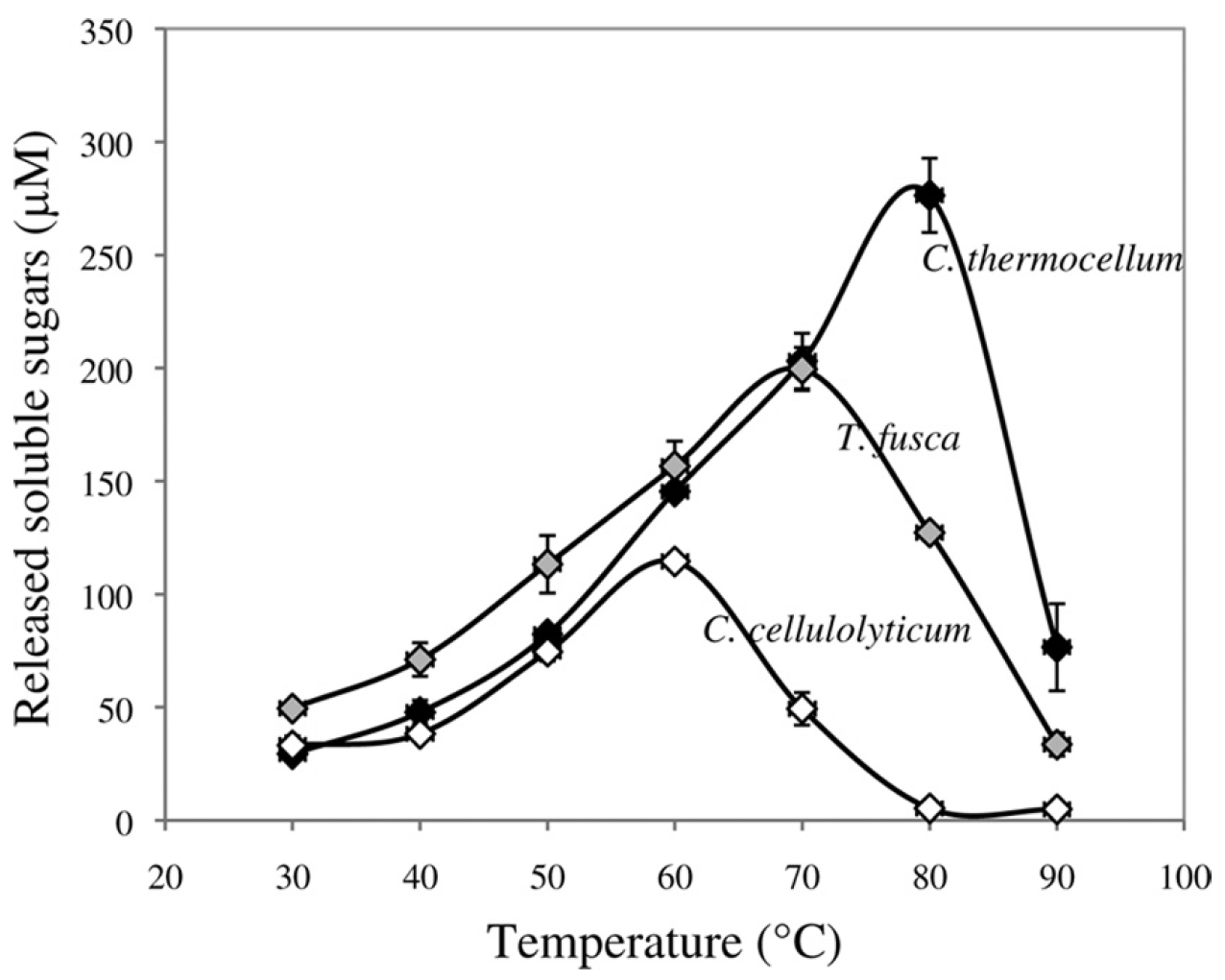 We studied three family 9 bacterial endoglucanases with optimum temperatures ranging from 60 C to 80 C to explore how increasing thermostability affects catalytic activity. We found that the most thermostable enzyme had the highest maximum activity at its optimum temperature, with no differences in activity at lower temperatures. In other words, at higher temperatures the thermostable homolog was a much better cellulase. This demonstrates why we are interested in discovering new highly active, highly stable enzymes.
We studied three family 9 bacterial endoglucanases with optimum temperatures ranging from 60 C to 80 C to explore how increasing thermostability affects catalytic activity. We found that the most thermostable enzyme had the highest maximum activity at its optimum temperature, with no differences in activity at lower temperatures. In other words, at higher temperatures the thermostable homolog was a much better cellulase. This demonstrates why we are interested in discovering new highly active, highly stable enzymes.
"Efficient Screening of Fungal Cellobiohydrolase Class I Enzymes for Thermostabilizing Sequence Blocks by SCHEMA Structure-Guided Recombination," P. Heinzelman, R. Komor, A. Kannan, P. Romero, X. Yu, S. Mohler, C. Snow, F. H. Arnold. Protein Engineering, Design and Selection 24, 1-10 (2010)
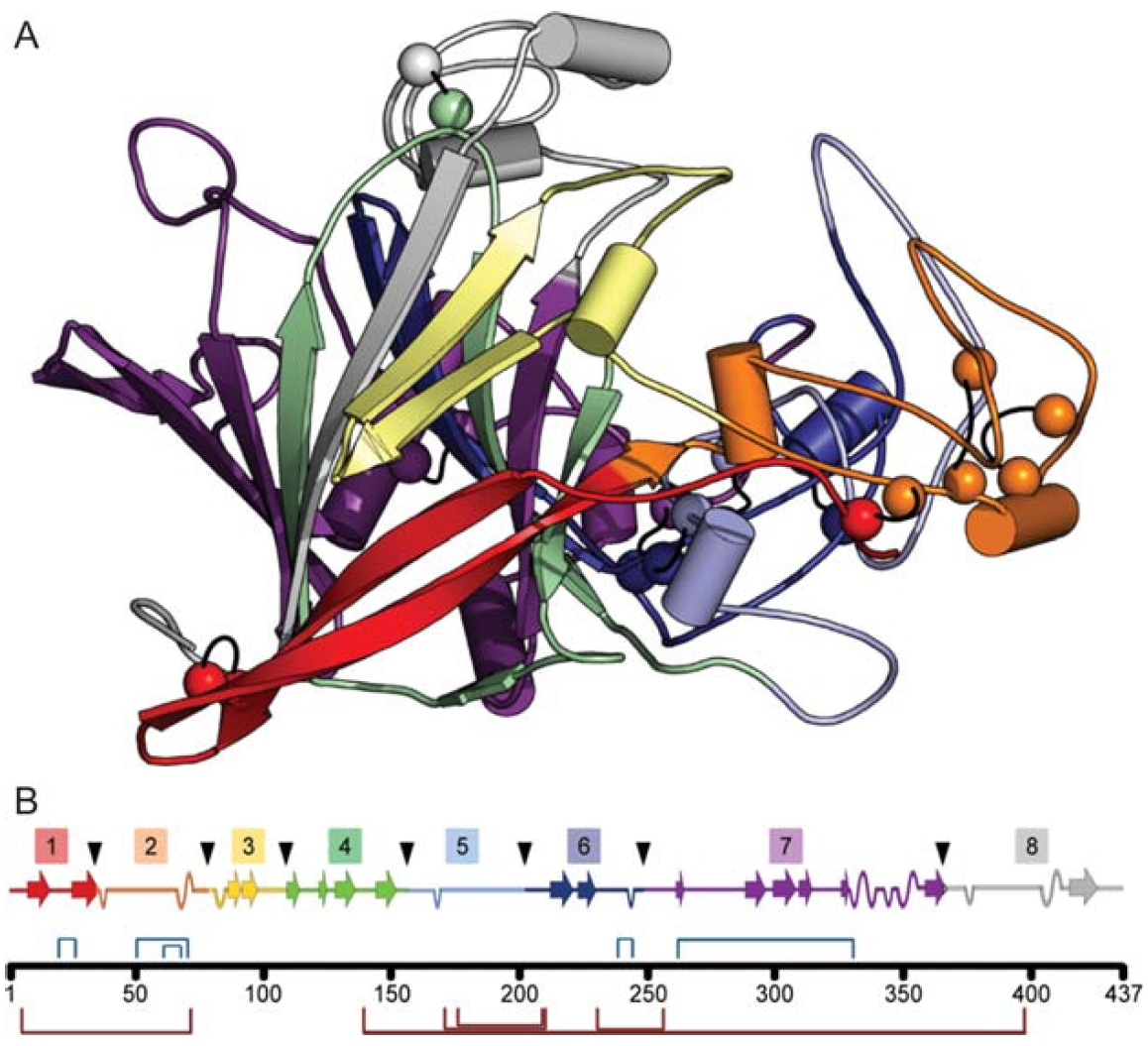 In this paper, we describe an efficient SCHEMA recombination approach to identifying stabilizing amino acid sequence blocks in a set of related enzymes. We determined the stability contributions of 32 protein blocks derived from 4 fungal cellobiohydrolase I’s (CBHIs or Cel7s) by substituting them individually into the most thermostable and highly expressed parent, CBH I from Talaromyces emersonii. Sixteen of 16 predicted thermostable chimeras, with an average of 37 mutations relative to the closest parent, were more thermostable than the most stable parent CBH I. The thermostable chimeras were also active on solid cellulose at temperatures at least 5ºC higher than the parent CBHIs.
In this paper, we describe an efficient SCHEMA recombination approach to identifying stabilizing amino acid sequence blocks in a set of related enzymes. We determined the stability contributions of 32 protein blocks derived from 4 fungal cellobiohydrolase I’s (CBHIs or Cel7s) by substituting them individually into the most thermostable and highly expressed parent, CBH I from Talaromyces emersonii. Sixteen of 16 predicted thermostable chimeras, with an average of 37 mutations relative to the closest parent, were more thermostable than the most stable parent CBH I. The thermostable chimeras were also active on solid cellulose at temperatures at least 5ºC higher than the parent CBHIs.
"A Family of Thermostable Fungal Cellulases Created by Structure-Guided Recombination," P. Heinzelman, C. D. Snow, I. Wu, C. Nguyen, A. Villalobos, S. Govindarajan, J. Minshull, F. H. Arnold. Proceedings of the National Academy of Sciences 106, 5610-5615 (2009).
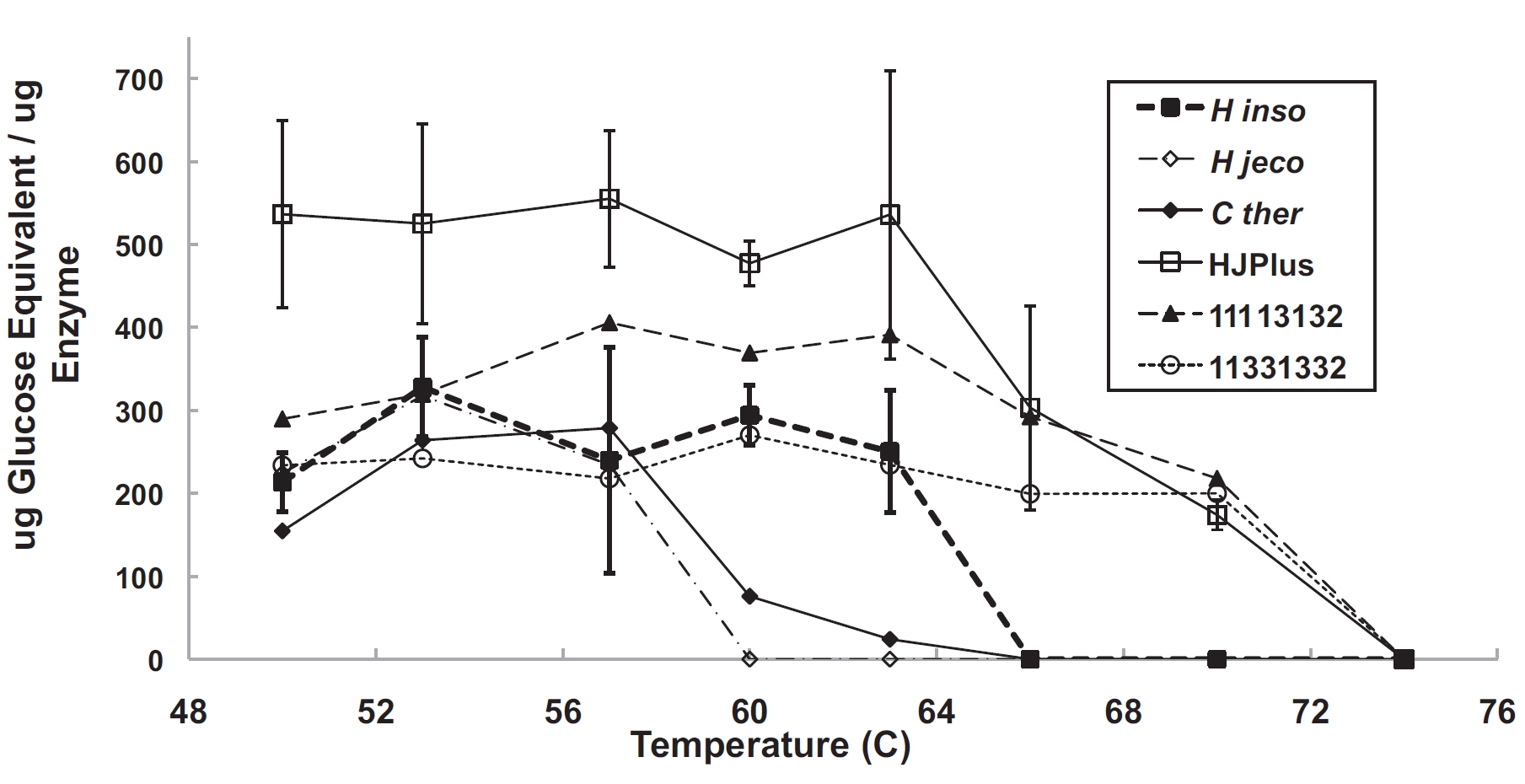 Here we constructed a chimeric library of fungal cellobiohydrolase II’s (CBHIIs or Cel6s) using SCHEMA recombination. We built linear regression models using the half-lives of 23 active chimeras and predicted thermostable chimera sequences. The resulting 15 thermostable CBH II enzymes differ from their closest natural homologs at up to 63 amino acid positions. Selected purified thermostable chimeras hydrolyzed phosphoric acid swollen cellulose at temperatures 7 to 15°C higher and hydrolyzed as much or more cellulose than the parent wild-type enzymes. This work demonstrates very well how quickly SCHEMA recombination can discovered highly stable, highly active enzymes.
Here we constructed a chimeric library of fungal cellobiohydrolase II’s (CBHIIs or Cel6s) using SCHEMA recombination. We built linear regression models using the half-lives of 23 active chimeras and predicted thermostable chimera sequences. The resulting 15 thermostable CBH II enzymes differ from their closest natural homologs at up to 63 amino acid positions. Selected purified thermostable chimeras hydrolyzed phosphoric acid swollen cellulose at temperatures 7 to 15°C higher and hydrolyzed as much or more cellulose than the parent wild-type enzymes. This work demonstrates very well how quickly SCHEMA recombination can discovered highly stable, highly active enzymes.
"SCHEMA Recombination of a Fungal Cellulase Uncovers a Single Mutation that Contributes Markedly to Stability." P. Heinzelman, C. D. Snow, M. A. Smith, X. Yu, K. Arvind, K. Bouleware, A. Villalobos, S. Govindarajan, J. Minshull, F.H. Arnold. Journal of Biological Chemistry 284, 26229-26233 (2009).
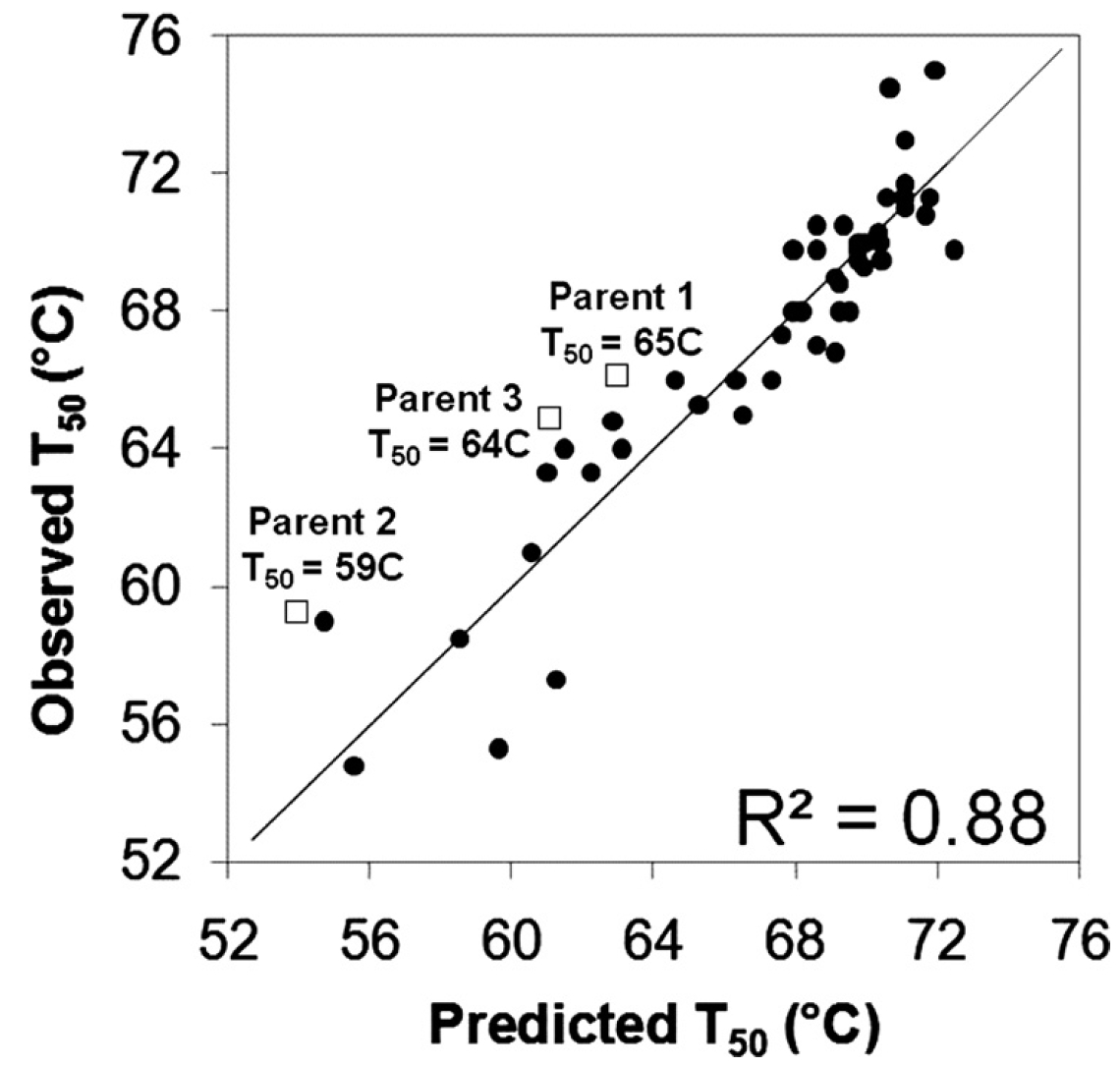 We refined the linear regression model for 54 characterized CBHII chimeras and identified a single block of sequence that adds 8.5 °C to thermostability. We then used site-directed mutagenesis to find that a single C313S substitution was responsible for the entire thermostabilizing effect. Introducing this mutation into selected wild-type CBHIIs decreased inactivation, increased maximum Avicel hydrolysis temperature, and improved long time hydrolysis performance. SCHEMA structure-guided recombination enabled quantitative prediction of cellulase chimera thermostability and efficient identification of stabilizing mutations.
We refined the linear regression model for 54 characterized CBHII chimeras and identified a single block of sequence that adds 8.5 °C to thermostability. We then used site-directed mutagenesis to find that a single C313S substitution was responsible for the entire thermostabilizing effect. Introducing this mutation into selected wild-type CBHIIs decreased inactivation, increased maximum Avicel hydrolysis temperature, and improved long time hydrolysis performance. SCHEMA structure-guided recombination enabled quantitative prediction of cellulase chimera thermostability and efficient identification of stabilizing mutations.

