Structure-guided protein recombination
 We are trying to understand the benefits of recombination (sex) in evolution. We also want to understand how to use it efficiently to make new proteins with new features and functions. Sex in the test tube is not limited to two parents, nor to sequences from the same species. We can recombine 32 parents. Or sequences from monkeys and worms. We want to understand the rules for molecular sex: how to do it, what it can make, and what we can learn from it. We have observed, for example, that sex in the test tube is an innovation generation machine.
We are trying to understand the benefits of recombination (sex) in evolution. We also want to understand how to use it efficiently to make new proteins with new features and functions. Sex in the test tube is not limited to two parents, nor to sequences from the same species. We can recombine 32 parents. Or sequences from monkeys and worms. We want to understand the rules for molecular sex: how to do it, what it can make, and what we can learn from it. We have observed, for example, that sex in the test tube is an innovation generation machine.
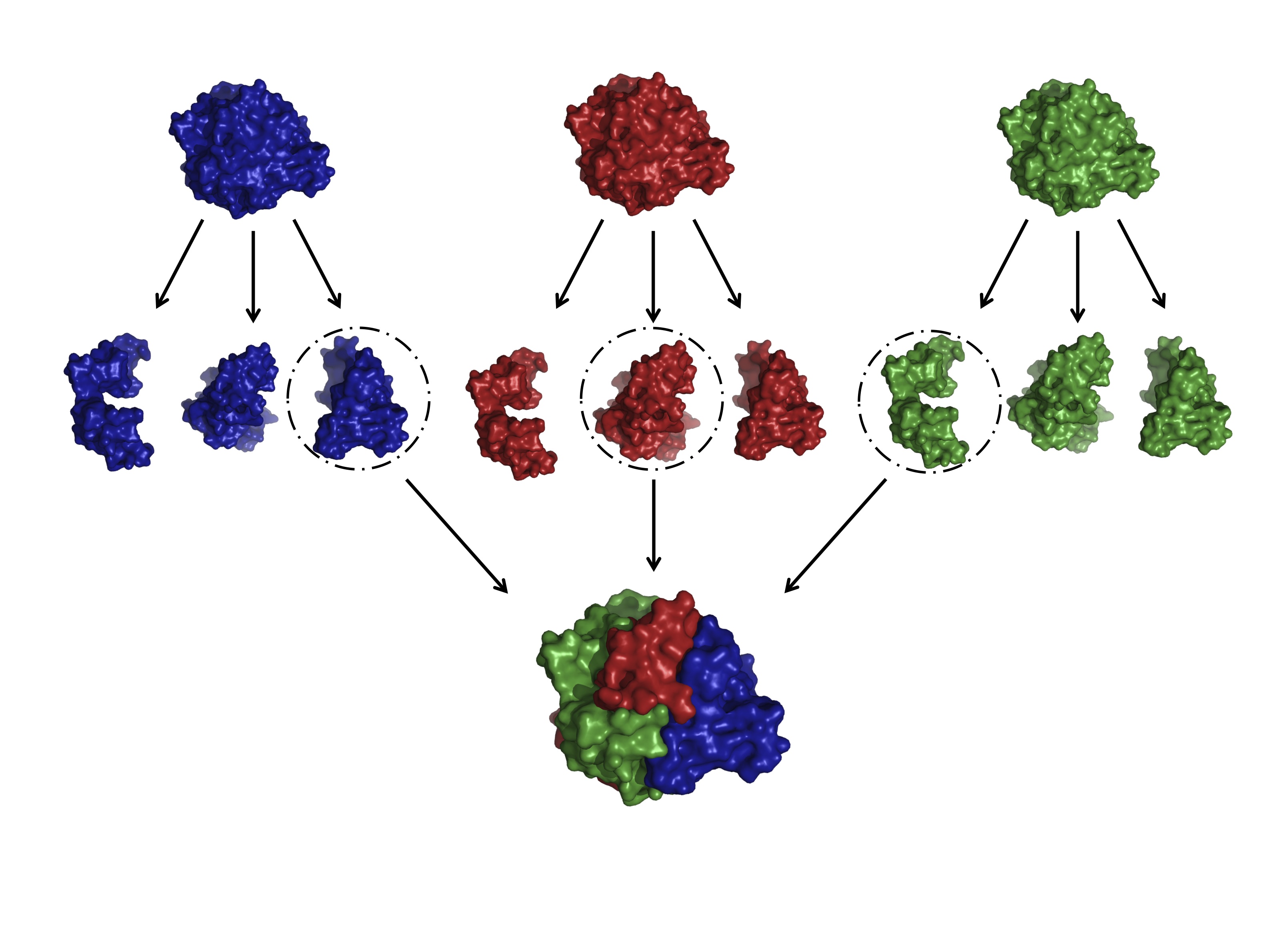
Homologous recombination is remarkably efficient for searching sequence space for functional proteins (i.e. it has a good chance of creating functional proteins) due to the conservative nature of homologous substitutions (they are less disruptive on average than random substitutions) and to the conservative nature of swapping blocks of sequence among related proteins. Chimeric proteins inherit the best and worst residues the parents have to offer, in new combinations that are not observed in nature. This leads to functional innovation.
We have developed computational tools that use protein structure information to design chimeric proteins and libraries of such proteins. These libraries are extremely diverse, with members that differ by tens or even hundreds of mutations while still maintaining a high proportion of sequences that fold and function. These chimeric proteins can be more stable than any of their parents. They can also catalyze reactions better than their parents, or even reactions their parents do not catalyze. We have also discovered that recombination leads to simplified (additive) sequence-function relationships that can be exploited to predict useful new sequences based on data from a small sampling of chimeras.
SCHEMA recombination
 Homologous recombination means swapping pieces of protein (blocks) between a set of homologs (parental proteins). The goal of site-directed SCHEMA recombination is to simultaneously maximize the mutation level of the chimeras and the probability the chimeric proteins will fold and function. We do this by minimizing the number of structural contacts that are disrupted when portions of sequence are inherited from different parent proteins. Using SCHEMA, we have made functional enzyme chimeras from parents sharing as little as 30% sequence identity. Guided by structural information, we have designed and constructed recombination libraries of a variety of enzymes, including beta-lactamases, arginases, cytochrome P450s, GH48 cellulases, GH6 cellulases, and GH7 cellulases.
Homologous recombination means swapping pieces of protein (blocks) between a set of homologs (parental proteins). The goal of site-directed SCHEMA recombination is to simultaneously maximize the mutation level of the chimeras and the probability the chimeric proteins will fold and function. We do this by minimizing the number of structural contacts that are disrupted when portions of sequence are inherited from different parent proteins. Using SCHEMA, we have made functional enzyme chimeras from parents sharing as little as 30% sequence identity. Guided by structural information, we have designed and constructed recombination libraries of a variety of enzymes, including beta-lactamases, arginases, cytochrome P450s, GH48 cellulases, GH6 cellulases, and GH7 cellulases.
We have discovered that the ‘recombination fitness landscape’ has a large additive component, which enables us to use simple linear regression models built from small data sets to predict highly stable chimera sequences. Homologous recombination thus gives us the opportunity to create and study a large number of functional enzymes whose properties vary significantly. With empirical models, we can accurately predict some of these properties and use these predictions to search for improved enzymes. We can also identify the sequence basis for variations in function.
Non-contiguous recombination
 We have extended our recombination design tools to include libraries where the blocks are not necessarily contiguous in the primary sequence. Although not contiguous along the polypeptide chain, the blocks are contiguous on the folded 3-D structure of the protein. Non-contiguous recombination further reduces structural disruption, as important contacts between residues not next to each other in the protein chain can be preserved. We expect that this will allow us to design chimeras and chimera libraries using more distantly-related parent proteins, further increasing the diversity of chimera progeny.
We have extended our recombination design tools to include libraries where the blocks are not necessarily contiguous in the primary sequence. Although not contiguous along the polypeptide chain, the blocks are contiguous on the folded 3-D structure of the protein. Non-contiguous recombination further reduces structural disruption, as important contacts between residues not next to each other in the protein chain can be preserved. We expect that this will allow us to design chimeras and chimera libraries using more distantly-related parent proteins, further increasing the diversity of chimera progeny.
Papers to get started:
"Structure-Guided Recombination Creates an Artificial Family of Cytochromes P450," C. R. Otey, M. Landwehr, J. B. Endelman, K. Hiraga, J. D. Bloom, F. H. Arnold. PLoS Biology 4(5):e112 (2006).
This paper describes a family of P450 enzymes we created using SCHEMA recombination. This library contains thousands of enzymes that are folded and active, with an average number of more than 70 amino acid mutations from the parents. Some of the enzymes were found to be more stable than any of the parental enzymes used for recombination. Some catalyze new reactions.
"A Diverse Family of Thermostable Cytochrome P450s Created By Recombination of Stabilizing Fragments," Y. Li, D. A. Drummond, A. M. Sawayama, C. D. Snow, J. D. Bloom, F. Arnold. Nature Biotechnology 25, 1051-1056 (2007).
Here we introduce an empirical model that can predict enzyme thermostability. By assuming that each piece of recombined protein (block) contributes additively to an enzyme's overall thermostability, we are able to accurately model the thermostabilities for a large number of P450s and successfully predict chimeric P450s that are much more stable than the parental enzymes used for recombination.
"A Family of Thermostable Fungal Cellulases Created by Structure-Guided Recombination," P. Heinzelman, C. D. Snow, I. Wu, C. Nguyen, A. Villalobos, S. Govindarajan, J. Minshull, F. H. Arnold. Proceedings of the National Academy of Sciences 106, 5610-5615 (2009).
Here we recombined three GH6 cellobiohydrolases to create 23 novel, active chimeric cellulases. Some of these cellulases were more stable than the most stable parental enzyme. Using sequence-stability data from these first 23 enzymes, 10 more active stable cellulases were made and these were up to 15°C more stable than any of the parents.
"SCHEMA-Designed Variants of Human Arginase I & II Reveal Sequence Elements Important to Stability and Catalysis," P. A. Romero, E. Stone, C. Lamb, L. Chantranupong, A. Krause, A. Miklos, R. A. Hughes, B. Fechtel, A. D. Ellington, F. H. Arnold, G. Georgiou. ACS Synthetic Biology 1, 221-228 (2012).
This paper models the long-term stabilities of a set of chimeric human arginases that might be used to treat cancer. Combining experimental measurements with sequence information, we discover a sequence-function relationship between an arginase's isoelectric point and its long-term stability.
"Random Field Model Reveals Structure of the Protein Recombinational Landscape,," P. A. Romero, F. H. Arnold. PLoS Computational Biology 8(10), e1002713 (2012). doi:10.1371/journal.pcbi.1002713
We have observed that the protein space accessible by homologous recombination is rich in functional sequences, and recombined fragments contribute additively to certain biophysical properties. In this paper we develop a random field model to explore these phenomena and to quantify the relative importance of sequence identity, crossover locations, and protein fold in producing functional chimeras. This model agrees quantitatively with results from numerous chimera experiments. It shows that the ‘recombination landscape’ is additive and explains why recombination makes new proteins that have many mutations but still fold and function.
"Chimeragenesis of distantly-related protein by noncontiguous recombination," M. A. Smith, P. A. Romero, T. Wu, E. M. Brustad, F. H. Arnold. Protein Science 22, 231-238 (2013).
We introduce a method for identifying elements of structure that can be shuffled among homologous proteins to create chimeras with less structural disruption. The x-ray crystal structure of a chimeric β-glucosidase that is half eukaryotic and half prokaryotic shows that the fragments maintain the backbone conformations found in their respective parental structures.
"Innovation by homologous recombination," D. L. Trudeau, M. A. Smith, F. H. Arnold. Current Opinion Chemical Biology 17, 902-909 (2013).
This review address covers methods to facilite recombination of homologous proteins and model sequence-function relationships. We give recent examples of chimeric proteins that have novel properties not exhibited by any of the parental proteins.

