Enzyme engineering to improve metabolic pathways
Sabine Brinkmann-Chen, Jackson Cahn, Mikiro Hayashi, Armin Baumschlager
New 'synthetic biology' or metabolic engineering approaches to producing fuels and chemicals from renewable resources require rerouting reactants in a metabolic network or the bottom-up integration of totally new pathways into existing ones. This means transplanting enzymes into biochemical contexts possibly very different from their native ones. And what is optimal for fuel production is hardly optimal for survival and fitness of the organism in which the enzymes evolved in nature. For example, a high titer of product is something that balanced metabolic enzyme networks usually try to evade. Many enzyme characteristics (e.g. stability, specific activity, cofactor usage, inhibition, or regulation) have to be specifically tailored to optimize metabolic flux toward the desired fuel or chemical.
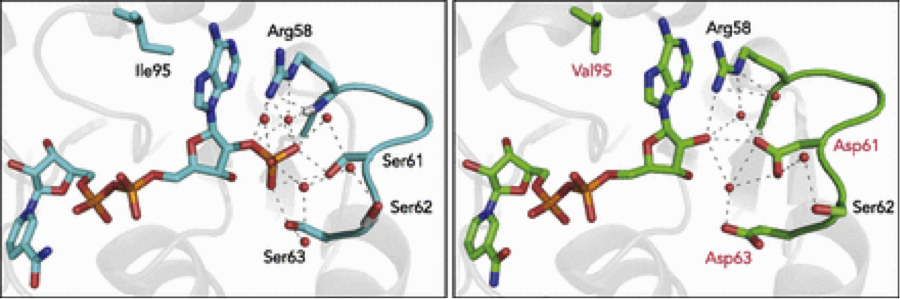
One particular problem our lab has focused on is nicotinamide cofactor usage. Cells use two distinct nicotinamide cofactors, NADH and NADPH, to donate and accept electrons for oxidative and reductive chemistry. Despite considerable structural similarity, most enzymes display exquisite specificity toward one or the other, allowing the cell to regulate whole sets of pathways by varying the relative rates of production of the cofactors. However, this regulation often runs counter to our goals as engineers, so the ability to interconvert enzymes from one specificity to another quickly and easily would be a powerful tool.
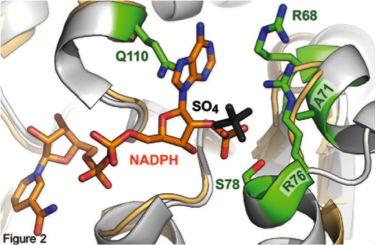 However, the phosphate which separates NADPH from NADH generally requires three or more side-chain interactions to bind (or not bind), making this a challenging problem for traditional single-mutation driven protein engineering techniques, such as site-saturation mutagenesis or error-prone PCR. For that reason, our current research focuses on developing structure- and sequence-based shortcuts to render this engineering task more straightforward. We have developed a simple recipe for the cofactor specificity reversal of ketol acid-reductoisomerases, and going forward hope to extend this to broader swathes of enzymes.
However, the phosphate which separates NADPH from NADH generally requires three or more side-chain interactions to bind (or not bind), making this a challenging problem for traditional single-mutation driven protein engineering techniques, such as site-saturation mutagenesis or error-prone PCR. For that reason, our current research focuses on developing structure- and sequence-based shortcuts to render this engineering task more straightforward. We have developed a simple recipe for the cofactor specificity reversal of ketol acid-reductoisomerases, and going forward hope to extend this to broader swathes of enzymes.
Recent papers:
"Engineered Ketol-Acid Reductoisomerase and Alcohol Dehydrogenase Enable Anaerobic 2-methylpropan-1-ol Production at Theoretical Yield in Escherichia coli S. Bastian, X. Li, J. T. Meyerowitz, C. D. Snow, M. M. Y. Chen, F. H. Arnold. Metabolic Engineering 13, 345-352 (2011).
The replacement of two enzymes in the NADPH-dependent isobutanol production pathway with NADH-dependent analogues enables anaerobic isobutanol production at 100% theoretical yield and at higher titer and productivity than the NADPH-dependent pathway.
"General Approach to Reversing Ketol-Acid Reductoisomerase Cofactor Dependence from NADPH to NADH S. Brinkmann-Chen, T. Flock, J. K. B. Cahn, C. D. Snow, E. M. Brustad, J. A. McIntosh, P. Meinhold, L. Zhang, F. H. Arnold. Proceedings of the National Academy of Sciences 110, 10946-10951 (2013). doi:10.1073/pnas.1306073110

